class: center, middle, title-slide # Skip your coffee break: Speed up MCMC convergence ### Daniel Turek & Olivier Gimenez ### last updated: 2022-06-24 --- ## Our `nimble` workflow so far 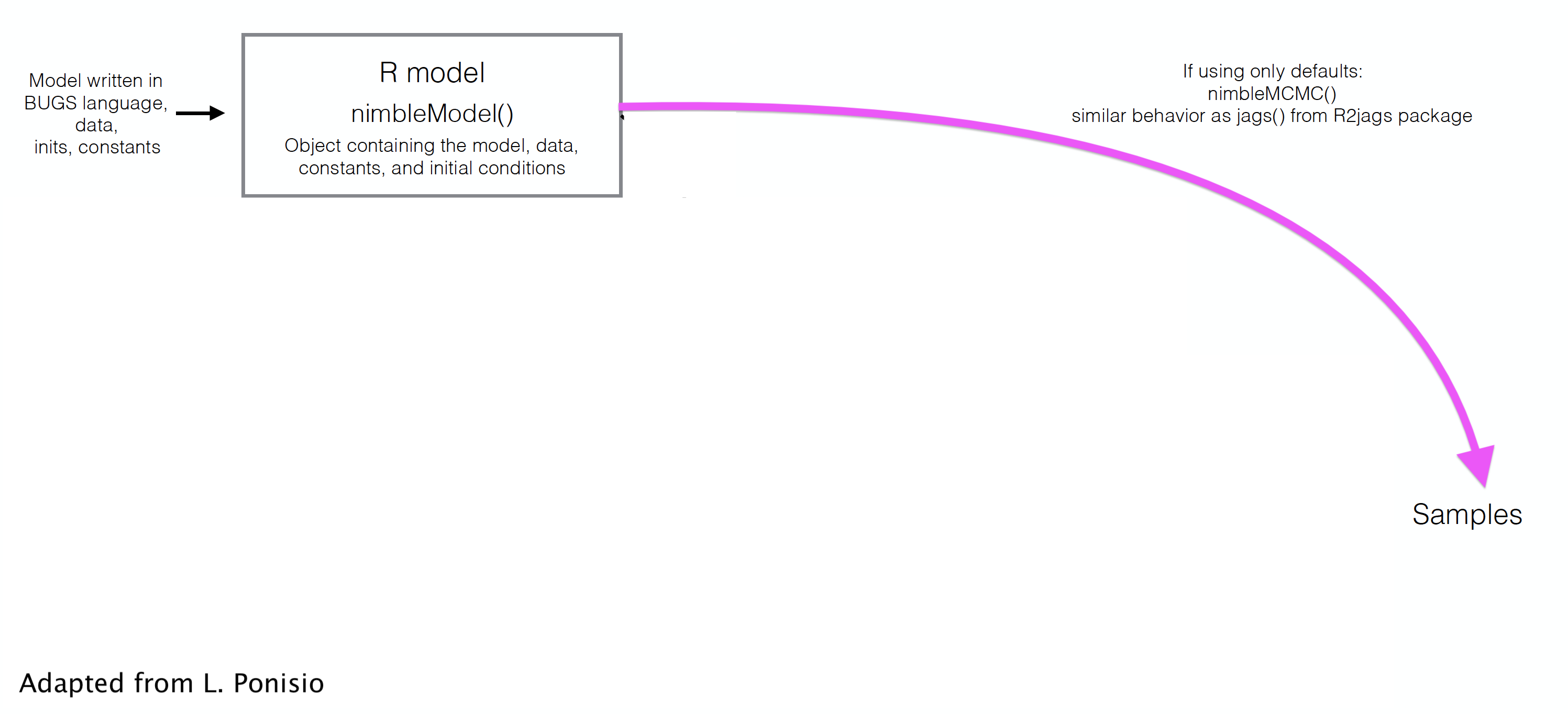 --- ## But `nimble` gives full access to the MCMC engine -- 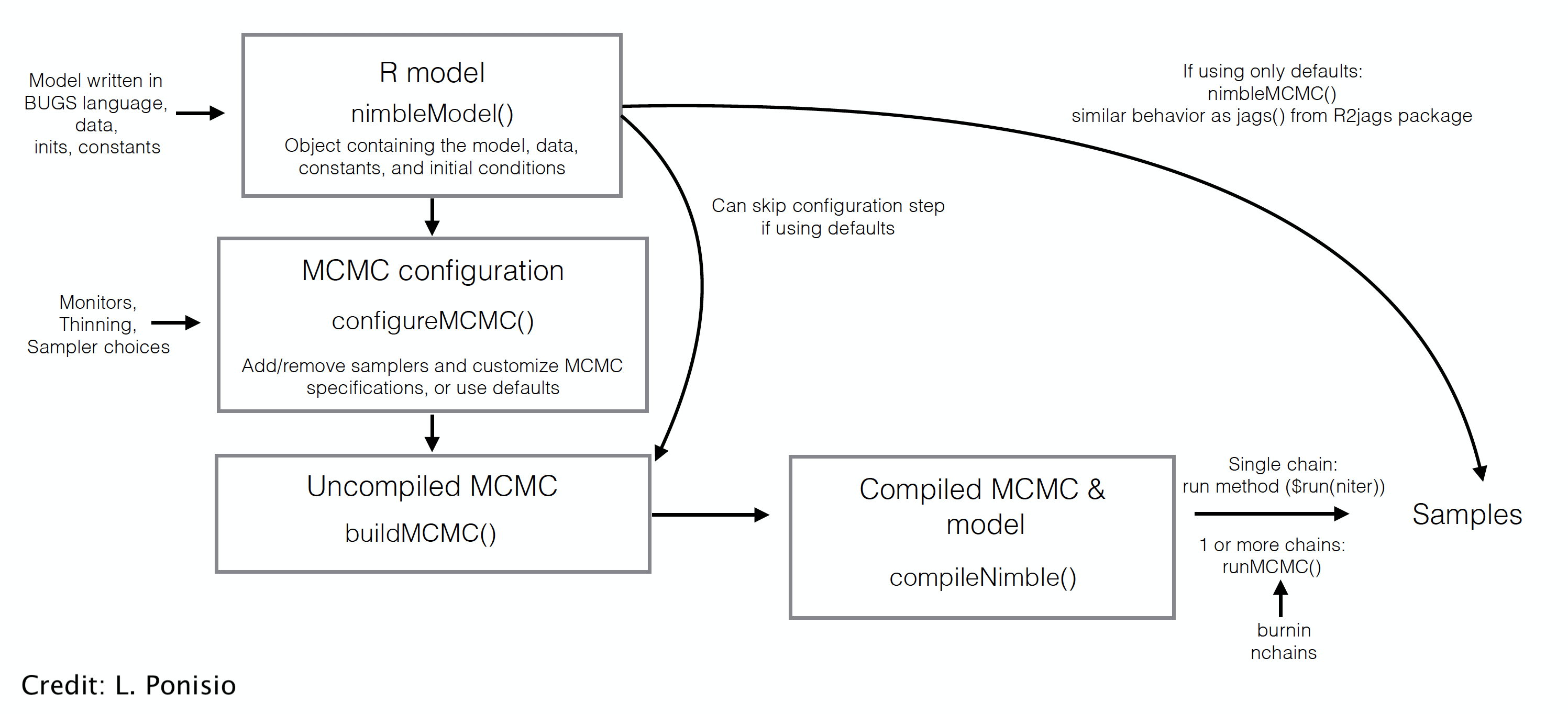 --- ## Steps to use NIMBLE at full capacity 1. Build the model. It is an R object. 2. Build the MCMC. 3. Compile the model and MCMC. 4. Run the MCMC. 5. Extract the samples. - `nimbleMCMC` does all of this at once. --- class: middle, center # Back to CJS models with Dipper data. --- ### Define model .tiny-font[ ```r hmm.phip <- nimbleCode({ delta[1] <- 1 # Pr(alive t = 1) = 1 delta[2] <- 0 # Pr(dead t = 1) = 0 phi ~ dunif(0, 1) # prior survival gamma[1,1] <- phi # Pr(alive t -> alive t+1) gamma[1,2] <- 1 - phi # Pr(alive t -> dead t+1) gamma[2,1] <- 0 # Pr(dead t -> alive t+1) gamma[2,2] <- 1 # Pr(dead t -> dead t+1) p ~ dunif(0, 1) # prior detection omega[1,1] <- 1 - p # Pr(alive t -> non-detected t) omega[1,2] <- p # Pr(alive t -> detected t) omega[2,1] <- 1 # Pr(dead t -> non-detected t) omega[2,2] <- 0 # Pr(dead t -> detected t) # likelihood for (i in 1:N){ z[i,first[i]] ~ dcat(delta[1:2]) for (j in (first[i]+1):T){ z[i,j] ~ dcat(gamma[z[i,j-1], 1:2]) y[i,j] ~ dcat(omega[z[i,j], 1:2]) } } }) ``` ] --- ### Run and summarise .tiny-font[ ```r mcmc.phip <- nimbleMCMC(code = hmm.phip, constants = my.constants, data = my.data, inits = initial.values, monitors = parameters.to.save, niter = n.iter, nburnin = n.burnin, nchains = n.chains) ``` ``` |-------------|-------------|-------------|-------------| |-------------------------------------------------------| |-------------|-------------|-------------|-------------| |-------------------------------------------------------| ``` ] .tiny-font[ ```r MCMCsummary(object = mcmc.phip, round = 2) ``` ``` mean sd 2.5% 50% 97.5% Rhat n.eff p 0.90 0.03 0.83 0.90 0.95 1.00 208 phi 0.56 0.03 0.51 0.56 0.61 1.01 489 ``` ] --- class: middle, center # Detailed Nimble workflow --- ## 1. Build the model (R object) .tiny-font[ ```r hmm.phip <- nimbleModel(code = hmm.phip, constants = my.constants, data = my.data, inits = initial.values()) ``` ``` defining model... ``` ``` building model... ``` ``` setting data and initial values... ``` ``` running calculate on model (any error reports that follow may simply reflect missing values in model variables) ... checking model sizes and dimensions... model building finished. ``` ] --- ## 2. Build the MCMC .small-font[ ```r phip.mcmc.configuration <- configureMCMC(hmm.phip) ``` ``` ===== Monitors ===== thin = 1: phi, p, z ===== Samplers ===== RW sampler (2) - phi - p posterior_predictive sampler (39) - z[] (39 elements) categorical sampler (1103) - z[] (1103 elements) ``` ```r phip.mcmc <- buildMCMC(phip.mcmc.configuration) ``` ] --- ## 3. Compile the model and MCMC .small-font[ ```r phip.model <- compileNimble(hmm.phip) ``` ``` compiling... this may take a minute. Use 'showCompilerOutput = TRUE' to see C++ compilation details. ``` ``` compilation finished. ``` ```r c.phip.mcmc <- compileNimble(phip.mcmc, project = phip.model) ``` ``` compiling... this may take a minute. Use 'showCompilerOutput = TRUE' to see C++ compilation details. compilation finished. ``` ] --- ## 4. Run the MCMC .small-font[ ```r samples <- runMCMC(c.phip.mcmc, niter = 1000) ``` ``` running chain 1... ``` ``` |-------------|-------------|-------------|-------------| |-------------------------------------------------------| ``` ```r # Alternative: # c.phip.mcmc$run(1000) # samples <- as.matrix(c.phip.mcmc$mvSamples) ``` ] --- ## 5. Look at results .small-font[ ```r summary(samples[,"phi"]) ``` ``` Min. 1st Qu. Median Mean 3rd Qu. Max. 0.5049 0.5452 0.5583 0.5755 0.5872 0.8214 ``` ```r summary(samples[,"p"]) ``` ``` Min. 1st Qu. Median Mean 3rd Qu. Max. 0.2289 0.8631 0.8929 0.8648 0.9171 0.9636 ``` ] --- class: middle, center # Why is it useful? --- ## Use and debug model in `R` + Makes your life easier when it comes to debugging + Inspect variables ```r hmm.phip$gamma ``` ``` [,1] [,2] [1,] 0.1071727 0.8928273 [2,] 0.0000000 1.0000000 ``` + Calculate likelihood ```r hmm.phip$calculate() ``` ``` [1] -2532.144 ``` --- ## Example of debugging a model in `R` + Pretend an impossible state was given in inits, making a dead bird alive again. .small-font[ ```r phip.model$calculate("z") # We can see there is a problem in z (states). ``` ``` [1] -Inf ``` ```r c(phip.model$calculate("z[5,]"), # Bird 5 is valid. phip.model$calculate("z[6,]")) # Bird 6 isn't. ``` ``` [1] -3.627276 -Inf ``` ```r phip.model$z[6,] # We have found the problem ``` ``` [1] 1 1 2 1 2 2 2 ``` ] --- ## Open the hood, and change/modify/write samplers + Slice samplers instead of Metropolis-Hastings. + Samplers on a log scale, especially for a variance, standard deviation, or precision parameter. + Blocking correlated parameters. + To know all samplers available in Nimble, type in `help(samplers)`. + Source code for samplers and distributions is **in R** and can be copied and modified. + Use [`compareMCMCs` package](https://github.com/nimble-dev/compareMCMCs) to compare options (including Stan and Jags!). --- class: middle, center ## Consider a model with wing length and individual random effect on survival. --- .tiny-font[ ```r hmm.phiwlrep <- nimbleCode({ p ~ dunif(0, 1) # prior detection omega[1,1] <- 1 - p # Pr(alive t -> non-detected t) omega[1,2] <- p # Pr(alive t -> detected t) omega[2,1] <- 1 # Pr(dead t -> non-detected t) omega[2,2] <- 0 # Pr(dead t -> detected t) for (i in 1:N){ * logit(phi[i]) <- beta[1] + beta[2] * winglength[i] + eps[i] * eps[i] ~ dnorm(mean = 0, sd = sdeps) gamma[1,1,i] <- phi[i] # Pr(alive t -> alive t+1) gamma[1,2,i] <- 1 - phi[i] # Pr(alive t -> dead t+1) gamma[2,1,i] <- 0 # Pr(dead t -> alive t+1) gamma[2,2,i] <- 1 # Pr(dead t -> dead t+1) } beta[1] ~ dnorm(mean = 0, sd = 1.5) beta[2] ~ dnorm(mean = 0, sd = 1.5) sdeps ~ dunif(0, 10) delta[1] <- 1 # Pr(alive t = 1) = 1 delta[2] <- 0 # Pr(dead t = 1) = 0 # likelihood for (i in 1:N){ z[i,first[i]] ~ dcat(delta[1:2]) for (j in (first[i]+1):T){ * z[i,j] ~ dcat(gamma[z[i,j-1], 1:2, i]) y[i,j] ~ dcat(omega[z[i,j], 1:2]) } } }) ``` ] --- ## Trace plot for standard deviation of the random effect (default sampler) <img src="4_speed_files/figure-html/unnamed-chunk-19-1.svg" style="display: block; margin: auto;" /> --- ## Change samplers + Good sampling strategies depend on the model and data. What are the samplers used by default? .tiny-font[ ```r mcmcConf <- configureMCMC(hmm.phiwlrep.m) ``` ``` ===== Monitors ===== thin = 1: p, beta, sdeps, z ===== Samplers ===== RW sampler (259) - p - beta[] (2 elements) - sdeps - eps[] (255 elements) posterior_predictive sampler (78) - eps[] (39 elements) - z[] (39 elements) categorical sampler (1103) - z[] (1103 elements) ``` ] --- ## Remove default sampler, and use slice sampler .tiny-font[ ```r mcmcConf$removeSamplers('sdeps') mcmcConf$addSampler(target = 'sdeps', * type = "slice") mcmcConf ``` ``` ===== Monitors ===== thin = 1: p, beta, sdeps, z ===== Samplers ===== slice sampler (1) - sdeps RW sampler (258) - p - beta[] (2 elements) - eps[] (255 elements) posterior_predictive sampler (78) - eps[] (39 elements) - z[] (39 elements) categorical sampler (1103) - z[] (1103 elements) ``` ] <br> <br> ``` |-------------|-------------|-------------|-------------| |-------------------------------------------------------| |-------------|-------------|-------------|-------------| |-------------------------------------------------------| ``` --- ## Trace plot for standard deviation of the random effect (slice sampler) <img src="4_speed_files/figure-html/unnamed-chunk-24-1.svg" style="display: block; margin: auto;" /> --- ## Which is better? + MCMC efficiency depends on both mixing and computation time. + MCMC efficiency = Effective Sample Size (ESS) / computation time. + MCMC efficiency is the number of effectively independent posterior samples generated per second. + ESS is different for each parameter. (Computation time is the same for each parameter.) + ESS can be estimated from packages `coda` or `mcmcse`. These give statistical estimates, so different runs will give different estimates. + Efficiency with default sampler = 25.7 / 21.53 = 1.19. + Efficiency with slice sampler = 21.25 / 30.37 = 0.7. --- ## Block sampling + High correlation in (regression) parameters may make independent samplers inefficient. <img src="4_speed_files/figure-html/unnamed-chunk-27-1.svg" style="display: block; margin: auto;" /> + Block sampling (propose candidate values from multivariate distribution) might help. --- ## Block sampling + Remove and replace independent RW samples by block sampling. Then proceed as usual. .tiny-font[ ```r mcmcConf$removeSamplers(c('beta[1]','beta[2]')) mcmcConf$addSampler(target = c('beta[1]','beta[2]'), * type = "RW_block") ``` ] --- ## Block sampling .tiny-font[ ```r mcmcConf ``` ``` ===== Monitors ===== thin = 1: p, beta, sdeps, z ===== Samplers ===== slice sampler (1) - sdeps RW_block sampler (1) - beta[1], beta[2] RW sampler (256) - p - eps[] (255 elements) posterior_predictive sampler (78) - eps[] (39 elements) - z[] (39 elements) categorical sampler (1103) - z[] (1103 elements) ``` ] --- ## Summary of strategies for improving MCMC -- + Choose better initial values. -- + Customize sampler choice (more in [Chapter 7 of the User's manual](https://r-nimble.org/html_manual/cha-mcmc.html)). -- + Reparameterize, e.g. standardize covariates, deal with parameter redundancy. -- + Rewrite the model. + Vectorize to improve computational efficiency (not covered). + Avoid long chains of deterministic dependencies. + Marginalize to remove parameters + Use new functions and new distributions written as nimbleFunctions. -- + Write new samplers that take advantage of particular model structures (not covered). -- + Using multiple cores with parallelization: see how-to at <https://r-nimble.org/nimbleExamples/parallelizing_NIMBLE.html> --- ## Marginalization + User-defined distributions is another neat feature of Nimble. + Integrate over latent states if those are not the focus of ecological inference (marginalization). + Marginalization often (but not always) improves MCMC. See [Ponisio et al. 2020](https://onlinelibrary.wiley.com/doi/full/10.1002/ece3.6053) for examples. + The [nimbleEcology](https://cran.r-project.org/web/packages/nimbleEcology/vignettes/Introduction_to_nimbleEcology.html) package implements capture-recapture models and HMMs with marginalization. --- ### Our model `\((\phi_A, \phi_B, \psi_{AB}, \psi_{BA}, p_A, p_B)\)` .small-font[ ```r multisite <- nimbleCode({ ... # Likelihood for (i in 1:N){ # Define latent state at first capture z[i,first[i]] <- y[i,first[i]] - 1 for (t in (first[i]+1):K){ # State process: draw S(t) given S(t-1) z[i,t] ~ dcat(gamma[z[i,t-1],1:3]) # Observation process: draw O(t) given S(t) y[i,t] ~ dcat(omega[z[i,t],1:3]) } } }) ``` ] --- ### Same model with nimbleEcology .tiny-font[ ```r multisite <- nimbleCode({ ... # initial state probs for(i in 1:N) { init[i, 1:4] <- gamma[ y[i, first[i] ] - 1, 1:4 ] # first state propagation } # likelihood for (i in 1:N){ y[i,(first[i]+1):K] ~ dHMM(init = init[i,1:4], # count data from first[i] + 1 probObs = omega[1:4,1:4], # observation matrix probTrans = gamma[1:4,1:4], # transition matrix len = K - first[i], # nb of occasions checkRowSums = 0) # do not check whether elements in a row sum tp 1 } ... ``` ] + This runs twice as fast as the standard formulation with explicit latent states. + Marginalizing typically gives better mixing. --- ### Reducing redundant calculations + So far, a row of the dataset is an individual. However, several individuals may share the same encounter history. -- + The contribution of `\(M\)` individuals with the same encounter history is the likelihood of this particular encounter history raised to the power `\(M\)`. -- + Using this so-called **weighted likelihood** greatly decreases the computational burden. -- + This idea is used in most computer programs that implement maximum likelihood. In the Bayesian framework, the same idea was proposed in [Turek et al. (2016)](https://doi.org/10.1007/s10651-016-0353-z). -- + Cannot be done in Jags. Can be done in nimble thanks to nimble functions! -- + The run is *much* faster. Also allows fitting models to big datasets. More details in dedicated Worksheet. --- background-color: #234f66 ## <span style="color:white">No live demo, but there is a worksheet.</span> .center[  ] --- ## Future directions for NIMBLE + NIMBLE is under active development. Contributors are welcome, including those who want to get involved but don't know where. + Faster building of models and algorithms. Ability to save and re-load compiled work. + Automatic differentiation of model calculations, enabling Hamiltonian Monte Carlo, other sampling strategies, and Laplace approximation. + Tools for building packages that use NIMBLE "under the hood". --- ## Further reading + Turek, D., de Valpine, P. & Paciorek, C.J. [Efficient Markov chain Monte Carlo sampling for hierarchical hidden Markov models](https://doi.org/10.1007/s10651-016-0353-z) *Environ Ecol Stat* 23: 549–564 (2016). + Ponisio, L.C., de Valpine, P., Michaud, N., and Turek, D. [One size does not fit all: Customizing MCMC methods for hierarchical models using NIMBLE](https://doi.org/10.1002/ece3.6053) *Ecol Evol.* 10: 2385–2416 (2020). + Nimble workshop to come 26-28 May, check out [here](https://r-nimble.org/nimble-virtual-short-course-may-26-28). + Nimble workshop material online available [here](https://github.com/nimble-training). + Nimble [manual](https://r-nimble.org/html_manual/cha-welcome-nimble.html) and [cheatsheet](https://r-nimble.org/cheatsheets/NimbleCheatSheet.pdf).