class: center, middle, title-slide # ‘Free the modeler in you’: Intro to Nimble ### The team (citation by Marc Kéry) ### last updated: 2021-05-17 --- # What is Nimble? <div class="figure" style="text-align: center"> <img src="img/ToddStudents_Meme.jpg" alt="(Meme created by Todd Arnold's wonderful students)" width="24%" /><img src="img/RobRob_Comment_edited.png" alt="(Meme created by Todd Arnold's wonderful students)" width="60%" /> <p class="caption">(Meme created by Todd Arnold's wonderful students)</p> </div> --- # What is Nimble? -- + **N**umerical **I**nference for statistical **M**odels using **B**ayesian and **L**ikelihood **E**stimation. -- + A framework for hierarchical statistical models and algorithms. -- + Uses almost the same model code as WinBUGS, OpenBUGS, and JAGS. -- + An extension of the BUGS language: additional syntax, custom functions and distributions. -- + A configurable system for MCMC. -- + A library of other methods (SMC, MCEM). ??? + Sequential Monte Carlo (particle filtering) + Monte Carlo Expectation Maximization (maximum likelihood) -- + A model-generic programming system to write new analysis methods. --- ## Load `nimble` package ```r library(nimble) ``` --- ## Build model, made of likelihood and priors ```r naive.survival.model <- nimbleCode({ # prior phi ~ dunif(0, 1) # likelihood y ~ dbinom(phi, n) }) ``` --- ## Syntax: what's new/better/different? + Vectorization ```r # JAGS (& Nimble) for(t in 1:Tmax){ x[t] <- Mu.x + epsilon[t] } # Nimble x[1:Tmax] <- Mu.x + epsilon[1:Tmax] ``` -- --- ## Syntax: what's new/better/different? + More flexible specification of distributions ```r # JAGS (& Nimble) for(t in 1:Tmax){ epsilon[t] ~ dnorm(0, tau) } tau <- pow(sigma, -2) sigma ~ dunif(0, 5) # Nimble for(t in 1:Tmax){ epsilon[t] ~ dnorm(0, sd = sigma) } sigma ~ dunif(0, 5) ``` --- ## Syntax: what's new/better/different? + Your own functions and distributions ```r x[1:Tmax] <- myNimbleFunction(a = Mu.x, b = epsilon[1:Tmax]) ``` ```r sigma ~ dCustomDistr(c = 0.5, z = 10) ``` --- ## Syntax: what's new/better/different? + The end of empty indices ```r # JAGS sum.x <- sum(x[]) # Nimble sum.x <- sum(x[1:Tmax]) ``` -- + & more... --- ## Read in data Back to our naive survival model: ```r naive.survival.model <- nimbleCode({ # prior phi ~ dunif(0, 1) # likelihood y ~ dbinom(phi, n) }) ``` ```r my.data <- list(n = 57, y = 19) ``` --- ## Distinguish constants and data To Nimble, not all "data" is data... ```r my.constants <- list(n = 57) my.data <- list(y = 19) ``` -- **Constants**: + Can never be changed + Must be provided when a model is defined (part of the model structure) + E.g. vector of known index values, variables used to define for-loops, etc. --- ## Distinguish constants and data To Nimble, not all "data" is data... ```r my.constants <- list(n = 57) my.data <- list(y = 19) ``` **Data**: + Can be changed without re-building the model + Can be (re-)simulated within a model + E.g. stuff that *only* appears to the left of a "~" -- For computational efficiency, better to specify as much as possible as constants. -- Nimble will help you with this! --- ## Specify initial values ```r initial.values <- function() list(phi = runif(1,0,1)) ``` -- ```r initial.values() ``` ``` $phi [1] 0.9287372 ``` --- ## Which parameters to save? ```r parameters.to.save <- c("phi") ``` --- ## MCMC details ```r n.iter <- 5000 n.burnin <- 1000 n.chains <- 2 n.thin <- 1 ``` -- Number of posterior samples per chain: `$$n.posterior = \frac{n.iter - n.burnin}{n.thin}$$` --- ## Run model, tadaa! ```r mcmc.output <- nimbleMCMC(code = naive.survival.model, data = my.data, constants = my.constants, inits = initial.values, monitors = parameters.to.save, thin = n.thin, niter = n.iter, nburnin = n.burnin, nchains = n.chains) ``` --- ## Explore MCMC outputs ```r str(mcmc.output) ``` ``` List of 2 $ chain1: num [1:4000, 1] 0.406 0.43 0.264 0.264 0.297 ... ..- attr(*, "dimnames")=List of 2 .. ..$ : NULL .. ..$ : chr "phi" $ chain2: num [1:4000, 1] 0.274 0.274 0.389 0.389 0.389 ... ..- attr(*, "dimnames")=List of 2 .. ..$ : NULL .. ..$ : chr "phi" ``` --- ## Explore MCMC outputs ```r head(mcmc.output$chain1) ``` ``` phi [1,] 0.4057891 [2,] 0.4297459 [3,] 0.2644534 [4,] 0.2644534 [5,] 0.2971877 [6,] 0.2971877 ``` --- ## Explore MCMC outputs .center.nogap[ <!-- --> ] --- ## Numerical summaries ```r library(MCMCvis) MCMCsummary(mcmc.output, round = 2) ``` ``` mean sd 2.5% 50% 97.5% Rhat n.eff phi 0.34 0.06 0.23 0.34 0.47 1 1793 ``` --- ## Trace and posterior density .pull-left[ ```r MCMCtrace(mcmc.output, pdf = FALSE) ``` ] -- .pull-right[ .center.nogap[ <!-- --> ] ] --- ## Trace and posterior density .pull-left[ ```r MCMCtrace(mcmc.output, pdf = FALSE, ind = TRUE, Rhat = TRUE, n.eff = TRUE) ``` ] -- .pull-right[ .center.nogap[ <!-- --> ] ] --- ## Our `nimble` workflow so far 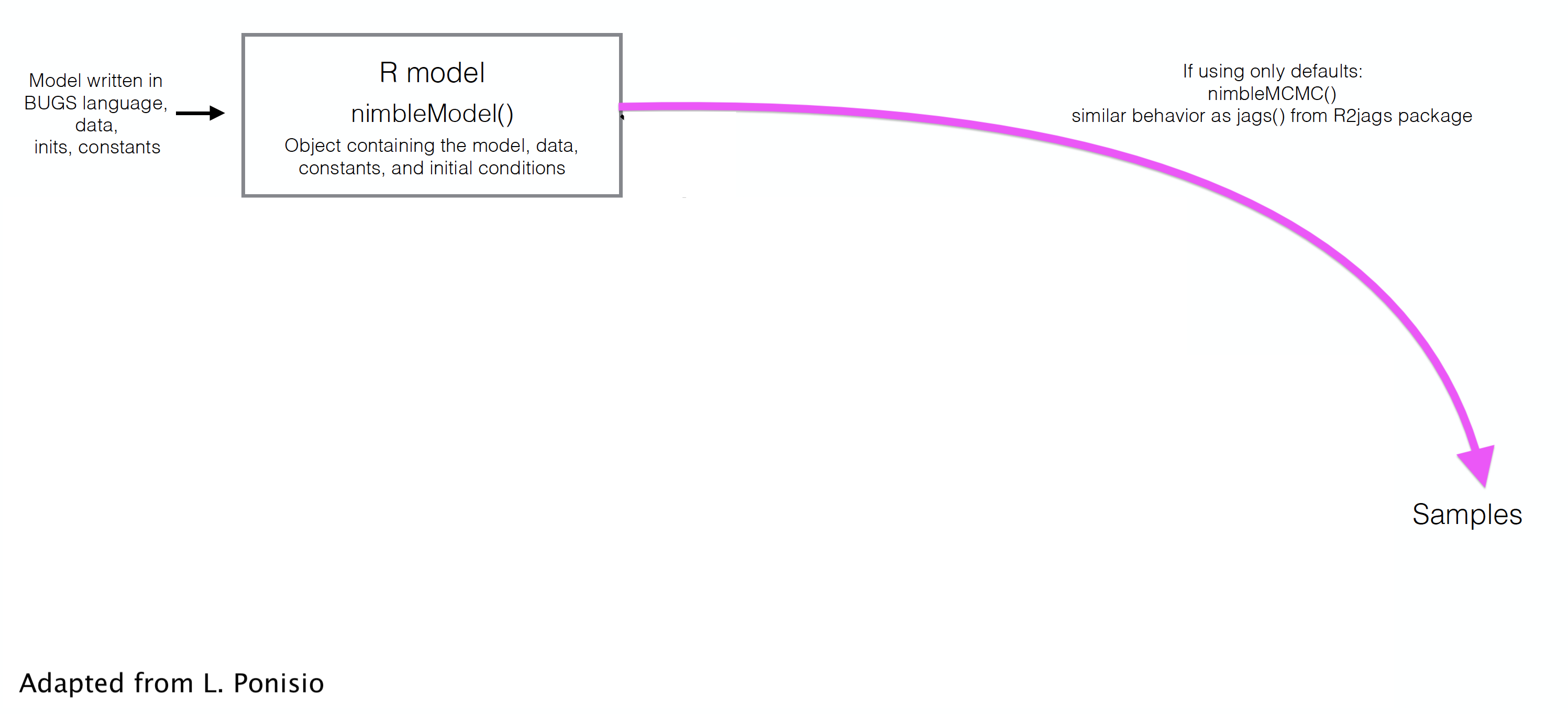 --- ## But `nimble` gives full access to the MCMC engine -- 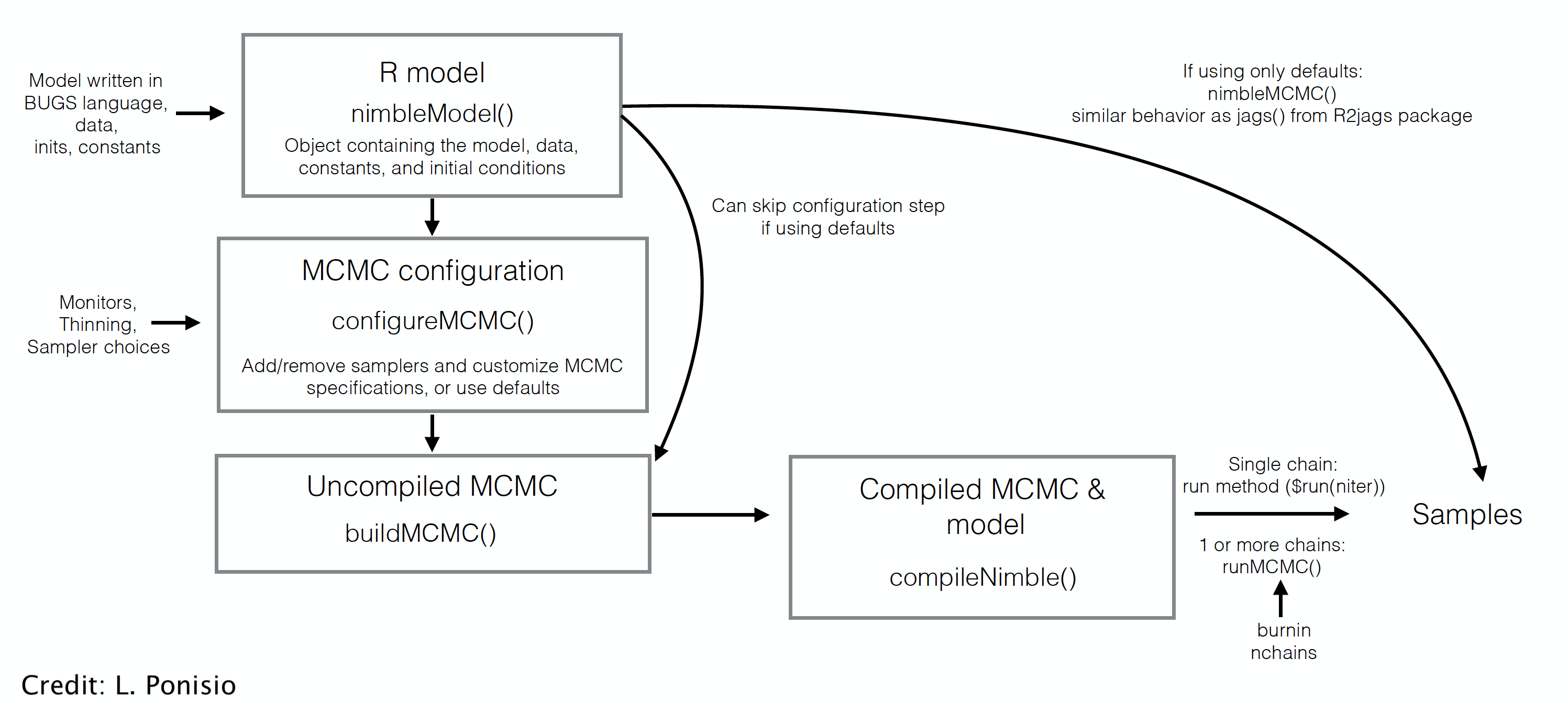 --- class: middle center background-color: black  --- ## Useful resources + Official website [https://r-nimble.org](https://r-nimble.org) + User Manual [https://r-nimble.org/html_manual/cha-welcome-nimble.html](https://r-nimble.org/html_manual/cha-welcome-nimble.html) and [cheatsheet](https://r-nimble.org/cheatsheets/NimbleCheatSheet.pdf). + Users mailing list [https://groups.google.com/forum/#!forum/nimble-users](https://groups.google.com/forum/#!forum/nimble-users) + Training material [https://github.com/nimble-training](https://github.com/nimble-training) + Reference to cite when using nimble in a publication: > de Valpine, P., D. Turek, C. J. Paciorek, C. Anderson-Bergman, D. Temple Lang, and R. Bodik (2017). [Programming With Models: Writing Statistical Algorithms for General Model Structures With NIMBLE](https://arxiv.org/pdf/1505.05093.pdf). *Journal of Computational and Graphical Statistics* **26** (2): 403–13. --- background-color: black # <span style="color:white">Live demo</span> <br> <br> .center[  ]